Temperature, Salinity, and Stratification¶
Now that we have learned the basic thermodynamics of seawater and understood how temperature and salinity combine to determine the stratifcation of water columns, we are ready to examine the global patterns of temperature, salinity and stratification. These patterns are fascinating and reveal much about the underlying ocean circulation. We will also learn how to identify distinct water masses both in geographic space and in temperature / salinity coordinates.
import xarray as xr
from matplotlib import pyplot as plt
import hvplot.xarray
import gsw
import numpy as np
import holoviews as hv
from IPython.display import Image
hv.extension('bokeh')
plt.rcParams['figure.figsize'] = (12,7)
Data Source: World Ocean Atlas¶
We will use data from the NOAA World Ocean Atlas, stored on the IRI Data Library at http://iridl.ldeo.columbia.edu/SOURCES/.NOAA/.NODC/.WOA09/.
World Ocean Atlas 2009 (WOA09) is a set of objectively analyzed (1° grid) climatological fields of in situ temperature, salinity, dissolved oxygen, Apparent Oxygen Utilization (AOU), percent oxygen saturation, phosphate, silicate, and nitrate at standard depth levels for annual, seasonal, and monthly compositing periods for the World Ocean. It also includes associated statistical fields of observed oceanographic profile data interpolated to standard depth levels on both 1° and 5° grids .
# basin masks
woa_mask = xr.open_dataset('http://iridl.ldeo.columbia.edu/SOURCES/.NOAA/.NODC/.WOA09/'
'.Masks/.basin/dods/').rename({'X':'lon', 'Y':'lat', 'Z': 'depth'})
basin_names = woa_mask.basin.attrs['CLIST'].split('\n')
basins_main = { n: basin_names[n-1] for n in [1,2,3,10] }
basins_marginal = { n: basin_names[n-1] for n in [4,5,6,7,11] }
woa_mask
<xarray.Dataset>
Dimensions: (depth: 33, lat: 180, lon: 360)
Coordinates:
* depth (depth) float32 0.0 10.0 20.0 30.0 ... 4e+03 4.5e+03 5e+03 5.5e+03
* lon (lon) float32 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5
* lat (lat) float32 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
Data variables:
basin (depth, lat, lon) float32 ...
Attributes:
Conventions: IRIDL- depth: 33
- lat: 180
- lon: 360
- depth(depth)float320.0 10.0 20.0 ... 5e+03 5.5e+03
- gridtype :
- 0
- units :
- m
array([ 0., 10., 20., 30., 50., 75., 100., 125., 150., 200., 250., 300., 400., 500., 600., 700., 800., 900., 1000., 1100., 1200., 1300., 1400., 1500., 1750., 2000., 2500., 3000., 3500., 4000., 4500., 5000., 5500.], dtype=float32) - lon(lon)float320.5 1.5 2.5 ... 357.5 358.5 359.5
- standard_name :
- longitude
- pointwidth :
- 1.0
- gridtype :
- 1
- units :
- degree_east
array([ 0.5, 1.5, 2.5, ..., 357.5, 358.5, 359.5], dtype=float32)
- lat(lat)float32-89.5 -88.5 -87.5 ... 88.5 89.5
- standard_name :
- latitude
- pointwidth :
- 1.0
- gridtype :
- 0
- units :
- degree_north
array([-89.5, -88.5, -87.5, -86.5, -85.5, -84.5, -83.5, -82.5, -81.5, -80.5, -79.5, -78.5, -77.5, -76.5, -75.5, -74.5, -73.5, -72.5, -71.5, -70.5, -69.5, -68.5, -67.5, -66.5, -65.5, -64.5, -63.5, -62.5, -61.5, -60.5, -59.5, -58.5, -57.5, -56.5, -55.5, -54.5, -53.5, -52.5, -51.5, -50.5, -49.5, -48.5, -47.5, -46.5, -45.5, -44.5, -43.5, -42.5, -41.5, -40.5, -39.5, -38.5, -37.5, -36.5, -35.5, -34.5, -33.5, -32.5, -31.5, -30.5, -29.5, -28.5, -27.5, -26.5, -25.5, -24.5, -23.5, -22.5, -21.5, -20.5, -19.5, -18.5, -17.5, -16.5, -15.5, -14.5, -13.5, -12.5, -11.5, -10.5, -9.5, -8.5, -7.5, -6.5, -5.5, -4.5, -3.5, -2.5, -1.5, -0.5, 0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, 10.5, 11.5, 12.5, 13.5, 14.5, 15.5, 16.5, 17.5, 18.5, 19.5, 20.5, 21.5, 22.5, 23.5, 24.5, 25.5, 26.5, 27.5, 28.5, 29.5, 30.5, 31.5, 32.5, 33.5, 34.5, 35.5, 36.5, 37.5, 38.5, 39.5, 40.5, 41.5, 42.5, 43.5, 44.5, 45.5, 46.5, 47.5, 48.5, 49.5, 50.5, 51.5, 52.5, 53.5, 54.5, 55.5, 56.5, 57.5, 58.5, 59.5, 60.5, 61.5, 62.5, 63.5, 64.5, 65.5, 66.5, 67.5, 68.5, 69.5, 70.5, 71.5, 72.5, 73.5, 74.5, 75.5, 76.5, 77.5, 78.5, 79.5, 80.5, 81.5, 82.5, 83.5, 84.5, 85.5, 86.5, 87.5, 88.5, 89.5], dtype=float32)
- basin(depth, lat, lon)float32...
- long_name :
- basin code
- units :
- ids
- scale_max :
- 58
- CLIST :
- Atlantic Ocean Pacific Ocean Indian Ocean Mediterranean Sea Baltic Sea Black Sea Red Sea Persian Gulf Hudson Bay Southern Ocean Arctic Ocean Sea of Japan Kara Sea Sulu Sea Baffin Bay East Mediterranean West Mediterranean Sea of Okhotsk Banda Sea Caribbean Sea Andaman Basin North Caribbean Gulf of Mexico Beaufort Sea South China Sea Barents Sea Celebes Sea Aleutian Basin Fiji Basin North American Basin West European Basin Southeast Indian Basin Coral Sea East Indian Basin Central Indian Basin Southwest Atlantic Basin Southeast Atlantic Basin Southeast Pacific Basin Guatemala Basin East Caroline Basin Marianas Basin Philippine Sea Arabian Sea Chile Basin Somali Basin Mascarene Basin Crozet Basin Guinea Basin Brazil Basin Argentine Basin Tasman Sea Atlantic Indian Basin Caspian Sea Sulu Sea II Venezuela Basin Bay of Bengal Java Sea East Indian Atlantic Basin
- valid_min :
- 1
- valid_max :
- 58
- scale_min :
- 1
[2138400 values with dtype=float32]
- Conventions :
- IRIDL
# actual data
url_base = 'http://iridl.ldeo.columbia.edu/SOURCES/.NOAA/.NODC/.WOA09/.Grid-1x1/.Annual'
url_temp = url_base + '/.temperature/.t_an/dods'
url_salt = url_base + '/.salinity/.s_an/dods '
woa_temp = xr.open_dataset(url_temp)
woa_temp_nobs = xr.open_dataarray(url_base + '/.temperature/.t_dd/dods')
woa_salt = xr.open_dataset(url_salt)
woa = xr.merge([woa_temp, woa_salt, woa_temp_nobs, woa_mask]).isel(time=0)
woa.load()
<xarray.Dataset>
Dimensions: (depth: 33, lat: 180, lon: 360)
Coordinates:
* lon (lon) float32 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5
* depth (depth) float32 0.0 10.0 20.0 30.0 ... 4e+03 4.5e+03 5e+03 5.5e+03
* lat (lat) float32 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
time datetime64[ns] 2008-01-01
Data variables:
t_an (depth, lat, lon) float32 nan nan nan nan nan ... nan nan nan nan
s_an (depth, lat, lon) float32 nan nan nan nan nan ... nan nan nan nan
t_dd (depth, lat, lon) float64 nan nan nan nan nan ... nan nan nan nan
basin (depth, lat, lon) float32 nan nan nan nan nan ... nan nan nan nan
Attributes:
Conventions: IRIDL- depth: 33
- lat: 180
- lon: 360
- lon(lon)float320.5 1.5 2.5 ... 357.5 358.5 359.5
- long_name :
- longitude
- pointwidth :
- 1.0
- bounds :
- lon_bnds
- axis :
- X
- modulus :
- 360.0
- description :
- Longitude Range: 0 to 360 degrees with 0 at Greenwich Meridian and increasing along the east.
- standard_name :
- longitude
- gridtype :
- 1
- units :
- degree_east
array([ 0.5, 1.5, 2.5, ..., 357.5, 358.5, 359.5], dtype=float32)
- depth(depth)float320.0 10.0 20.0 ... 5e+03 5.5e+03
- standard_name :
- depth
- long_name :
- depth
- positive :
- down
- axis :
- Z
- description :
- Standard Depth Levels
- gridtype :
- 0
- units :
- m
array([ 0., 10., 20., 30., 50., 75., 100., 125., 150., 200., 250., 300., 400., 500., 600., 700., 800., 900., 1000., 1100., 1200., 1300., 1400., 1500., 1750., 2000., 2500., 3000., 3500., 4000., 4500., 5000., 5500.], dtype=float32) - lat(lat)float32-89.5 -88.5 -87.5 ... 88.5 89.5
- long_name :
- latitude
- pointwidth :
- 1.0
- bounds :
- lat_bnds
- axis :
- Y
- description :
- Latitude Range: -90 to 90 degrees with 0 at the Equator and increasing along the north.
- standard_name :
- latitude
- gridtype :
- 0
- units :
- degree_north
array([-89.5, -88.5, -87.5, -86.5, -85.5, -84.5, -83.5, -82.5, -81.5, -80.5, -79.5, -78.5, -77.5, -76.5, -75.5, -74.5, -73.5, -72.5, -71.5, -70.5, -69.5, -68.5, -67.5, -66.5, -65.5, -64.5, -63.5, -62.5, -61.5, -60.5, -59.5, -58.5, -57.5, -56.5, -55.5, -54.5, -53.5, -52.5, -51.5, -50.5, -49.5, -48.5, -47.5, -46.5, -45.5, -44.5, -43.5, -42.5, -41.5, -40.5, -39.5, -38.5, -37.5, -36.5, -35.5, -34.5, -33.5, -32.5, -31.5, -30.5, -29.5, -28.5, -27.5, -26.5, -25.5, -24.5, -23.5, -22.5, -21.5, -20.5, -19.5, -18.5, -17.5, -16.5, -15.5, -14.5, -13.5, -12.5, -11.5, -10.5, -9.5, -8.5, -7.5, -6.5, -5.5, -4.5, -3.5, -2.5, -1.5, -0.5, 0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, 10.5, 11.5, 12.5, 13.5, 14.5, 15.5, 16.5, 17.5, 18.5, 19.5, 20.5, 21.5, 22.5, 23.5, 24.5, 25.5, 26.5, 27.5, 28.5, 29.5, 30.5, 31.5, 32.5, 33.5, 34.5, 35.5, 36.5, 37.5, 38.5, 39.5, 40.5, 41.5, 42.5, 43.5, 44.5, 45.5, 46.5, 47.5, 48.5, 49.5, 50.5, 51.5, 52.5, 53.5, 54.5, 55.5, 56.5, 57.5, 58.5, 59.5, 60.5, 61.5, 62.5, 63.5, 64.5, 65.5, 66.5, 67.5, 68.5, 69.5, 70.5, 71.5, 72.5, 73.5, 74.5, 75.5, 76.5, 77.5, 78.5, 79.5, 80.5, 81.5, 82.5, 83.5, 84.5, 85.5, 86.5, 87.5, 88.5, 89.5], dtype=float32) - time()datetime64[ns]2008-01-01
- pointwidth :
- 0
- gridtype :
- 0
array('2008-01-01T00:00:00.000000000', dtype='datetime64[ns]')
- t_an(depth, lat, lon)float32nan nan nan nan ... nan nan nan nan
- grid_mapping :
- crs
- standard_name :
- sea_water_temperature
- cell_methods :
- time:mean within years time:mean over years
- long_name :
- Sea Water Temperature
- units :
- degrees Celsius
- description :
- The objectively interpolated mean field for Sea Water Temperature for annual time period at each one-degree square at each standard depth.
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [-1.6027, -1.583 , -1.5641, ..., -1.6467, -1.6357, -1.6209], [-1.6168, -1.605 , -1.5937, ..., -1.6421, -1.6363, -1.6276], [-1.6125, -1.6064, -1.6008, ..., -1.6203, -1.6188, -1.6154]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [-1.8245, -1.8032, -1.7863, ..., -1.855 , -1.8493, -1.8386], [-1.8001, -1.7884, -1.7767, ..., -1.8201, -1.8158, -1.8093], [-1.7704, -1.7638, -1.756 , ..., -1.7801, -1.7798, -1.7754]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., ... [-0.6856, -0.6856, -0.6856, ..., -0.6856, -0.6856, -0.6856], [-0.6856, -0.6856, -0.6856, ..., -0.6856, -0.6856, -0.6856], [-0.6856, -0.6856, -0.6856, ..., -0.6856, -0.6856, -0.6856]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - s_an(depth, lat, lon)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- sea_water_salinity
- cell_methods :
- time:mean within years time:mean over years
- salinity_scale :
- Practical Salinity Scale(PSS-78)
- long_name :
- Sea Water Salinity
- units :
- unitless
- grid_mapping :
- crs
- description :
- The objectively interpolated mean field for Sea Water Salinity for annual time period at each one-degree square at each standard depth.
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [31.8196, 31.8635, 31.884 , ..., 31.6614, 31.7311, 31.7661], [31.5965, 31.6369, 31.6655, ..., 31.5004, 31.5093, 31.5742], [31.395 , 31.4156, 31.4342, ..., 31.3159, 31.3376, 31.3604]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [32.4103, 32.4583, 32.4824, ..., 32.2429, 32.2778, 32.3209], [32.2732, 32.3243, 32.3533, ..., 32.1821, 32.1926, 32.259 ], [32.1368, 32.142 , 32.1698, ..., 32.0648, 32.0723, 32.105 ]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., ... [34.9423, 34.9423, 34.9423, ..., 34.9423, 34.9423, 34.9423], [34.9423, 34.9423, 34.9423, ..., 34.9423, 34.9423, 34.9423], [34.9423, 34.9423, 34.9423, ..., 34.9423, 34.9423, 34.9423]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - t_dd(depth, lat, lon)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Number of Observations
- units :
- unitless
- grid_mapping :
- crs
- description :
- The number of observations of Sea Water Temperature in each one-degree square of the World Ocean at each standard depth level
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 2., 2., 1.], [ 1., 1., 0., ..., 0., 0., 0.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 1., 2., 2., ..., 0., 0., 0.], [ 1., 0., 0., ..., 2., 2., 2.], [ 1., 1., 0., ..., 0., 0., 0.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - basin(depth, lat, lon)float32nan nan nan nan ... nan nan nan nan
- long_name :
- basin code
- units :
- ids
- scale_max :
- 58
- CLIST :
- Atlantic Ocean Pacific Ocean Indian Ocean Mediterranean Sea Baltic Sea Black Sea Red Sea Persian Gulf Hudson Bay Southern Ocean Arctic Ocean Sea of Japan Kara Sea Sulu Sea Baffin Bay East Mediterranean West Mediterranean Sea of Okhotsk Banda Sea Caribbean Sea Andaman Basin North Caribbean Gulf of Mexico Beaufort Sea South China Sea Barents Sea Celebes Sea Aleutian Basin Fiji Basin North American Basin West European Basin Southeast Indian Basin Coral Sea East Indian Basin Central Indian Basin Southwest Atlantic Basin Southeast Atlantic Basin Southeast Pacific Basin Guatemala Basin East Caroline Basin Marianas Basin Philippine Sea Arabian Sea Chile Basin Somali Basin Mascarene Basin Crozet Basin Guinea Basin Brazil Basin Argentine Basin Tasman Sea Atlantic Indian Basin Caspian Sea Sulu Sea II Venezuela Basin Bay of Bengal Java Sea East Indian Atlantic Basin
- valid_min :
- 1
- valid_max :
- 58
- scale_min :
- 1
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]], dtype=float32)
- Conventions :
- IRIDL
We will now augment the raw dataset with derived quantities, including pressure (calculated from depth), absolute salinity, conservative temperature, and potential density at various reference levels.
woa['pres'] = gsw.p_from_z(-woa.depth, 0)
woa['s_a'] = gsw.SA_from_SP(woa.s_an, woa.pres, woa.lon, woa.lat)
woa['c_t'] = gsw.CT_from_t(woa.s_a, woa.t_an, woa.pres)
woa['sigma_0'] = gsw.sigma0(woa.s_a, woa.c_t)
woa['sigma_2'] = gsw.sigma2(woa.s_a, woa.c_t)
woa['sigma_4'] = gsw.sigma4(woa.s_a, woa.c_t)
woa
<xarray.Dataset>
Dimensions: (depth: 33, lat: 180, lon: 360)
Coordinates:
* lon (lon) float32 0.5 1.5 2.5 3.5 4.5 ... 355.5 356.5 357.5 358.5 359.5
* depth (depth) float32 0.0 10.0 20.0 30.0 ... 4e+03 4.5e+03 5e+03 5.5e+03
* lat (lat) float32 -89.5 -88.5 -87.5 -86.5 -85.5 ... 86.5 87.5 88.5 89.5
time datetime64[ns] 2008-01-01
Data variables:
t_an (depth, lat, lon) float32 nan nan nan nan nan ... nan nan nan nan
s_an (depth, lat, lon) float32 nan nan nan nan nan ... nan nan nan nan
t_dd (depth, lat, lon) float64 nan nan nan nan nan ... nan nan nan nan
basin (depth, lat, lon) float32 nan nan nan nan nan ... nan nan nan nan
pres (depth) float64 0.0 10.06 20.11 ... 4.573e+03 5.087e+03 5.602e+03
s_a (depth, lat, lon) float64 nan nan nan nan nan ... nan nan nan nan
c_t (depth, lat, lon) float64 nan nan nan nan nan ... nan nan nan nan
sigma_0 (depth, lat, lon) float64 nan nan nan nan nan ... nan nan nan nan
sigma_2 (depth, lat, lon) float64 nan nan nan nan nan ... nan nan nan nan
sigma_4 (depth, lat, lon) float64 nan nan nan nan nan ... nan nan nan nan
Attributes:
Conventions: IRIDL- depth: 33
- lat: 180
- lon: 360
- lon(lon)float320.5 1.5 2.5 ... 357.5 358.5 359.5
- long_name :
- longitude
- pointwidth :
- 1.0
- bounds :
- lon_bnds
- axis :
- X
- modulus :
- 360.0
- description :
- Longitude Range: 0 to 360 degrees with 0 at Greenwich Meridian and increasing along the east.
- standard_name :
- longitude
- gridtype :
- 1
- units :
- degree_east
array([ 0.5, 1.5, 2.5, ..., 357.5, 358.5, 359.5], dtype=float32)
- depth(depth)float320.0 10.0 20.0 ... 5e+03 5.5e+03
- standard_name :
- depth
- long_name :
- depth
- positive :
- down
- axis :
- Z
- description :
- Standard Depth Levels
- gridtype :
- 0
- units :
- m
array([ 0., 10., 20., 30., 50., 75., 100., 125., 150., 200., 250., 300., 400., 500., 600., 700., 800., 900., 1000., 1100., 1200., 1300., 1400., 1500., 1750., 2000., 2500., 3000., 3500., 4000., 4500., 5000., 5500.], dtype=float32) - lat(lat)float32-89.5 -88.5 -87.5 ... 88.5 89.5
- long_name :
- latitude
- pointwidth :
- 1.0
- bounds :
- lat_bnds
- axis :
- Y
- description :
- Latitude Range: -90 to 90 degrees with 0 at the Equator and increasing along the north.
- standard_name :
- latitude
- gridtype :
- 0
- units :
- degree_north
array([-89.5, -88.5, -87.5, -86.5, -85.5, -84.5, -83.5, -82.5, -81.5, -80.5, -79.5, -78.5, -77.5, -76.5, -75.5, -74.5, -73.5, -72.5, -71.5, -70.5, -69.5, -68.5, -67.5, -66.5, -65.5, -64.5, -63.5, -62.5, -61.5, -60.5, -59.5, -58.5, -57.5, -56.5, -55.5, -54.5, -53.5, -52.5, -51.5, -50.5, -49.5, -48.5, -47.5, -46.5, -45.5, -44.5, -43.5, -42.5, -41.5, -40.5, -39.5, -38.5, -37.5, -36.5, -35.5, -34.5, -33.5, -32.5, -31.5, -30.5, -29.5, -28.5, -27.5, -26.5, -25.5, -24.5, -23.5, -22.5, -21.5, -20.5, -19.5, -18.5, -17.5, -16.5, -15.5, -14.5, -13.5, -12.5, -11.5, -10.5, -9.5, -8.5, -7.5, -6.5, -5.5, -4.5, -3.5, -2.5, -1.5, -0.5, 0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, 10.5, 11.5, 12.5, 13.5, 14.5, 15.5, 16.5, 17.5, 18.5, 19.5, 20.5, 21.5, 22.5, 23.5, 24.5, 25.5, 26.5, 27.5, 28.5, 29.5, 30.5, 31.5, 32.5, 33.5, 34.5, 35.5, 36.5, 37.5, 38.5, 39.5, 40.5, 41.5, 42.5, 43.5, 44.5, 45.5, 46.5, 47.5, 48.5, 49.5, 50.5, 51.5, 52.5, 53.5, 54.5, 55.5, 56.5, 57.5, 58.5, 59.5, 60.5, 61.5, 62.5, 63.5, 64.5, 65.5, 66.5, 67.5, 68.5, 69.5, 70.5, 71.5, 72.5, 73.5, 74.5, 75.5, 76.5, 77.5, 78.5, 79.5, 80.5, 81.5, 82.5, 83.5, 84.5, 85.5, 86.5, 87.5, 88.5, 89.5], dtype=float32) - time()datetime64[ns]2008-01-01
- pointwidth :
- 0
- gridtype :
- 0
array('2008-01-01T00:00:00.000000000', dtype='datetime64[ns]')
- t_an(depth, lat, lon)float32nan nan nan nan ... nan nan nan nan
- grid_mapping :
- crs
- standard_name :
- sea_water_temperature
- cell_methods :
- time:mean within years time:mean over years
- long_name :
- Sea Water Temperature
- units :
- degrees Celsius
- description :
- The objectively interpolated mean field for Sea Water Temperature for annual time period at each one-degree square at each standard depth.
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [-1.6027, -1.583 , -1.5641, ..., -1.6467, -1.6357, -1.6209], [-1.6168, -1.605 , -1.5937, ..., -1.6421, -1.6363, -1.6276], [-1.6125, -1.6064, -1.6008, ..., -1.6203, -1.6188, -1.6154]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [-1.8245, -1.8032, -1.7863, ..., -1.855 , -1.8493, -1.8386], [-1.8001, -1.7884, -1.7767, ..., -1.8201, -1.8158, -1.8093], [-1.7704, -1.7638, -1.756 , ..., -1.7801, -1.7798, -1.7754]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., ... [-0.6856, -0.6856, -0.6856, ..., -0.6856, -0.6856, -0.6856], [-0.6856, -0.6856, -0.6856, ..., -0.6856, -0.6856, -0.6856], [-0.6856, -0.6856, -0.6856, ..., -0.6856, -0.6856, -0.6856]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - s_an(depth, lat, lon)float32nan nan nan nan ... nan nan nan nan
- standard_name :
- sea_water_salinity
- cell_methods :
- time:mean within years time:mean over years
- salinity_scale :
- Practical Salinity Scale(PSS-78)
- long_name :
- Sea Water Salinity
- units :
- unitless
- grid_mapping :
- crs
- description :
- The objectively interpolated mean field for Sea Water Salinity for annual time period at each one-degree square at each standard depth.
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [31.8196, 31.8635, 31.884 , ..., 31.6614, 31.7311, 31.7661], [31.5965, 31.6369, 31.6655, ..., 31.5004, 31.5093, 31.5742], [31.395 , 31.4156, 31.4342, ..., 31.3159, 31.3376, 31.3604]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [32.4103, 32.4583, 32.4824, ..., 32.2429, 32.2778, 32.3209], [32.2732, 32.3243, 32.3533, ..., 32.1821, 32.1926, 32.259 ], [32.1368, 32.142 , 32.1698, ..., 32.0648, 32.0723, 32.105 ]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., ... [34.9423, 34.9423, 34.9423, ..., 34.9423, 34.9423, 34.9423], [34.9423, 34.9423, 34.9423, ..., 34.9423, 34.9423, 34.9423], [34.9423, 34.9423, 34.9423, ..., 34.9423, 34.9423, 34.9423]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - t_dd(depth, lat, lon)float64nan nan nan nan ... nan nan nan nan
- long_name :
- Number of Observations
- units :
- unitless
- grid_mapping :
- crs
- description :
- The number of observations of Sea Water Temperature in each one-degree square of the World Ocean at each standard depth level
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 2., 2., 1.], [ 1., 1., 0., ..., 0., 0., 0.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 1., 2., 2., ..., 0., 0., 0.], [ 1., 0., 0., ..., 2., 2., 2.], [ 1., 1., 0., ..., 0., 0., 0.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.], [ 0., 0., 0., ..., 0., 0., 0.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]]) - basin(depth, lat, lon)float32nan nan nan nan ... nan nan nan nan
- long_name :
- basin code
- units :
- ids
- scale_max :
- 58
- CLIST :
- Atlantic Ocean Pacific Ocean Indian Ocean Mediterranean Sea Baltic Sea Black Sea Red Sea Persian Gulf Hudson Bay Southern Ocean Arctic Ocean Sea of Japan Kara Sea Sulu Sea Baffin Bay East Mediterranean West Mediterranean Sea of Okhotsk Banda Sea Caribbean Sea Andaman Basin North Caribbean Gulf of Mexico Beaufort Sea South China Sea Barents Sea Celebes Sea Aleutian Basin Fiji Basin North American Basin West European Basin Southeast Indian Basin Coral Sea East Indian Basin Central Indian Basin Southwest Atlantic Basin Southeast Atlantic Basin Southeast Pacific Basin Guatemala Basin East Caroline Basin Marianas Basin Philippine Sea Arabian Sea Chile Basin Somali Basin Mascarene Basin Crozet Basin Guinea Basin Brazil Basin Argentine Basin Tasman Sea Atlantic Indian Basin Caspian Sea Sulu Sea II Venezuela Basin Bay of Bengal Java Sea East Indian Atlantic Basin
- valid_min :
- 1
- valid_max :
- 58
- scale_min :
- 1
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., ... ..., [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.], [11., 11., 11., ..., 11., 11., 11.]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - pres(depth)float640.0 10.06 ... 5.087e+03 5.602e+03
- standard_name :
- depth
- long_name :
- depth
- positive :
- down
- axis :
- Z
- description :
- Standard Depth Levels
- gridtype :
- 0
- units :
- m
array([ 0. , 10.0554684 , 20.11142784, 30.16787822, 50.28225153, 75.42797832, 100.57677078, 125.72862766, 150.8835477 , 201.2025722 , 251.5338342 , 301.87732362, 402.60094429, 503.37335332, 604.19446971, 705.06421233, 805.98249995, 906.94925122, 1007.96438468, 1109.02781877, 1210.13947182, 1311.29926206, 1412.50710765, 1513.76292662, 1767.11181903, 2020.75874803, 2528.94156481, 3038.301051 , 3548.8268504 , 4060.5085888 , 4573.33588663, 5087.29837213, 5602.38569503]) - s_a(depth, lat, lon)float64nan nan nan nan ... nan nan nan nan
- standard_name :
- sea_water_salinity
- cell_methods :
- time:mean within years time:mean over years
- salinity_scale :
- Practical Salinity Scale(PSS-78)
- long_name :
- Sea Water Salinity
- units :
- unitless
- grid_mapping :
- crs
- description :
- The objectively interpolated mean field for Sea Water Salinity for annual time period at each one-degree square at each standard depth.
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [31.97240237, 32.01648388, 32.03705172, ..., 31.81353415, 31.88353903, 31.91867671], [31.74825288, 31.78882175, 31.81753506, ..., 31.65176273, 31.66068159, 31.7258692 ], [31.54580566, 31.56648522, 31.58515625, ..., 31.46637793, 31.48816508, 31.51105612]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - c_t(depth, lat, lon)float64nan nan nan nan ... nan nan nan nan
- standard_name :
- sea_water_salinity
- cell_methods :
- time:mean within years time:mean over years
- salinity_scale :
- Practical Salinity Scale(PSS-78)
- long_name :
- Sea Water Salinity
- units :
- unitless
- grid_mapping :
- crs
- description :
- The objectively interpolated mean field for Sea Water Salinity for annual time period at each one-degree square at each standard depth.
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [-1.59548214, -1.57576602, -1.55682601, ..., -1.63946344, -1.62849936, -1.61368486], [-1.60937544, -1.59757559, -1.58626558, ..., -1.63467168, -1.62885754, -1.62019321], [-1.60483559, -1.59873232, -1.59312938, ..., -1.61258496, -1.61110129, -1.60771077]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - sigma_0(depth, lat, lon)float64nan nan nan nan ... nan nan nan nan
- standard_name :
- sea_water_salinity
- cell_methods :
- time:mean within years time:mean over years
- salinity_scale :
- Practical Salinity Scale(PSS-78)
- long_name :
- Sea Water Salinity
- units :
- unitless
- grid_mapping :
- crs
- description :
- The objectively interpolated mean field for Sea Water Salinity for annual time period at each one-degree square at each standard depth.
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [25.59790153, 25.63306036, 25.64923885, ..., 25.47053499, 25.52684798, 25.55490941], [25.41717289, 25.44968552, 25.47262952, ..., 25.33976692, 25.34685117, 25.39932252], [25.25357471, 25.27014956, 25.28511148, ..., 25.1895845 , 25.20715062, 25.2255691 ]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - sigma_2(depth, lat, lon)float64nan nan nan nan ... nan nan nan nan
- standard_name :
- sea_water_salinity
- cell_methods :
- time:mean within years time:mean over years
- salinity_scale :
- Practical Salinity Scale(PSS-78)
- long_name :
- Sea Water Salinity
- units :
- unitless
- grid_mapping :
- crs
- description :
- The objectively interpolated mean field for Sea Water Salinity for annual time period at each one-degree square at each standard depth.
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [35.06460844, 35.09773888, 35.11235726, ..., 34.94284954, 34.99722256, 35.0237221 ], [34.88874663, 34.91979128, 34.9415098 , ..., 34.81465928, 34.82121664, 34.87197891], [34.72846576, 34.74428627, 34.75856141, ..., 34.66638059, 34.68346473, 34.70126092]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]]) - sigma_4(depth, lat, lon)float64nan nan nan nan ... nan nan nan nan
- standard_name :
- sea_water_salinity
- cell_methods :
- time:mean within years time:mean over years
- salinity_scale :
- Practical Salinity Scale(PSS-78)
- long_name :
- Sea Water Salinity
- units :
- unitless
- grid_mapping :
- crs
- description :
- The objectively interpolated mean field for Sea Water Salinity for annual time period at each one-degree square at each standard depth.
array([[[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [44.10303826, 44.13426281, 44.14741725, ..., 43.98655414, 44.03910007, 44.06413173], [43.93176702, 43.9614308 , 43.98199736, ..., 43.86080232, 43.86686485, 43.91601663], [43.77462375, 43.78973456, 43.8033636 , ..., 43.71433523, 43.73096453, 43.7481743 ]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]])
- Conventions :
- IRIDL
Before diving into the actual data, let’s first examine the number of observations in each grid cell. This shows how some parts of the ocean are much better sampled than others.
woa.t_dd.where(woa.t_dd > 0).hvplot('lon', 'lat', logz=True, width=700, height=400)
Later we will split up the ocean by basin. Here is how WOA defines the different basins.
woa.basin.where(woa.basin<20).hvplot('lon', 'lat', logz=True, width=700, height=400, cmap='Category20')
print(woa.basin.attrs['CLIST'])
Atlantic Ocean
Pacific Ocean
Indian Ocean
Mediterranean Sea
Baltic Sea
Black Sea
Red Sea
Persian Gulf
Hudson Bay
Southern Ocean
Arctic Ocean
Sea of Japan
Kara Sea
Sulu Sea
Baffin Bay
East Mediterranean
West Mediterranean
Sea of Okhotsk
Banda Sea
Caribbean Sea
Andaman Basin
North Caribbean
Gulf of Mexico
Beaufort Sea
South China Sea
Barents Sea
Celebes Sea
Aleutian Basin
Fiji Basin
North American Basin
West European Basin
Southeast Indian Basin
Coral Sea
East Indian Basin
Central Indian Basin
Southwest Atlantic Basin
Southeast Atlantic Basin
Southeast Pacific Basin
Guatemala Basin
East Caroline Basin
Marianas Basin
Philippine Sea
Arabian Sea
Chile Basin
Somali Basin
Mascarene Basin
Crozet Basin
Guinea Basin
Brazil Basin
Argentine Basin
Tasman Sea
Atlantic Indian Basin
Caspian Sea
Sulu Sea II
Venezuela Basin
Bay of Bengal
Java Sea
East Indian Atlantic Basin
3D Temperature¶
def map_section_plot(data, label, data_range, levels, cmap='viridis'):
ds = hv.Dataset(data)
img = ds.to(hv.Image, ['lon', 'lat'], label=label)
img = img.redim.range(**{data.name: data_range})
posx = hv.streams.PointerX()
vline = hv.DynamicMap(lambda x: hv.VLine(x or 180.5), streams=[posx])
lats = np.arange(-90, 91)
def cross_section(x):
mesh = hv.QuadMesh((lats, woa.depth, data.sel(lon=x if x else 180, method='nearest').data[:-1]),
kdims=['lat', 'depth'], label=(label + ' Section'))
contours = hv.operation.contours(mesh, levels=levels,
overlaid=False, filled=True)
return contours
crosssection = hv.DynamicMap(cross_section, streams=[posx])
layout = (img * vline) + crosssection
# set view options
dict_spec = {'Image': {'plot': dict(colorbar=True, toolbar='above', width=600),
'style': dict(cmap=cmap)},
'NdOverlay': {'plot': dict(bgcolor='grey', invert_yaxis=True, width=600, height=200)},
'Layout': {'plot': dict(show_title=False)},
'Polygons': {'plot': dict(colorbar=True, invert_yaxis=True, width=600),
'style': dict(cmap=cmap, line_width=0.1)}
}
layout.cols(1)
layout = layout.opts(dict_spec)
return layout
map_section_plot(woa.t_an, 'Temperature', (-2,30), np.arange(-2,31), cmap='RdBu_r')
Observations¶
The warmest water is near the surface in the tropical regions, forming a bowl-shaped lens in the upper ~800 meters. This is called the thermocline.
There are clear zonal asymmetries in surface temperature. The western boundaries of the basins tend to be warmer, with colder water near the eastern boundaries.
There is something special going on at the equator, especially in the Pacific. It’s relatively cold, and the thermocline is shallow.
Below 1000 m, the water is mostly very cold (< 5 C).
In the polar regions, the coldest water is actually found near the surface! There is a mid-depth temperature maximum.
3D Salinity¶
map_section_plot(woa.s_an, 'Salinity', (32,38), np.arange(32,38.1, 0.25))
Observations¶
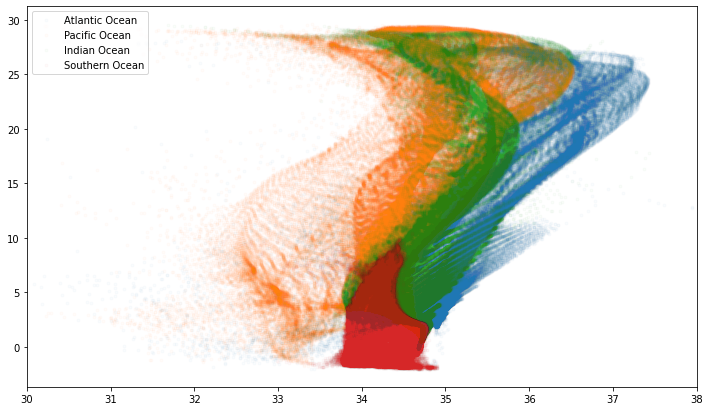
Salinity has a more complex structure than temperature.
The Atlantic is overall saltier than the Pacific.
The saltiest parts of the oceans are the extra-tropical-latitudes.
The equator is relatively fresh near the surface.
Salinity sections across the Atlantic reveal the interleaving of several different water masses including
Mediterranean Water
North Atlantic Deep Water
Antarctic Intermediate Water
Antarctic Bottom Water
3D Potential Density¶
map_section_plot(woa.sigma_0, 'Potential Density (sigma0)', (22.,28.), np.arange(22,28.1, 0.25))
map_section_plot(woa.sigma_4, 'Potential Density (sigma4)', (40.,46.5), np.arange(40,46.5, 0.25))
Observations¶
Potential density reflects the mechanical stratification of the ocean most closely of any of the quantities we have examined so far.
For the most part, potential density is monotonically increasing with depth.
BUT there are exceptions. Each potential density variable shows density “inversions” far from its reference level.
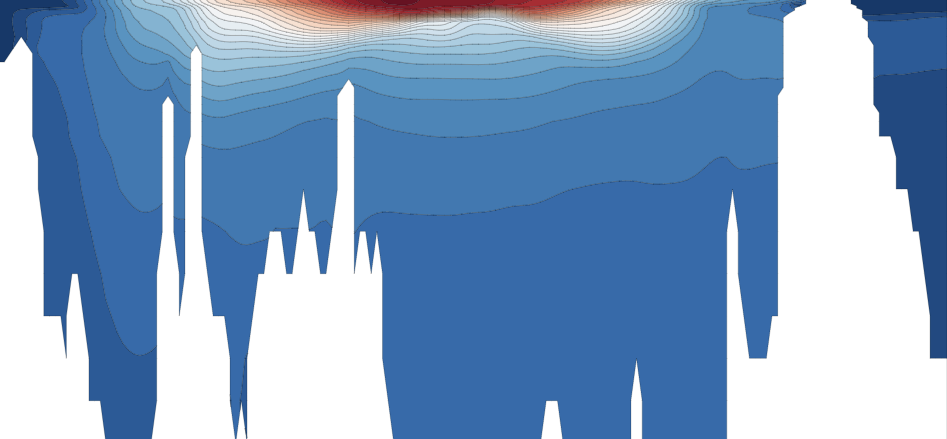
Potential density surface approximately represent pathways that water parcels can move along without experiencing a restoring force. Therefore, they serve as connections between the surface and deep ocean.
These connections are strongest in the Southern Ocean region.
Image('02_images/neutral_plane.png')
woa['thick'] = woa.depth.diff(dim='depth')
woa['thick'][0] = 5
# add an area element
earth_radius = 6.371e6
tot_area = (4 * np.pi * earth_radius**2)
woa['area'] = earth_radius**2 * np.radians(1.) * np.radians(1.) * np.cos(np.radians(woa.lat))
woa['vol'] = woa.thick * woa.area
vol_full, _ = xr.broadcast(woa.vol, woa.lon)
fig, (ax1, ax2) = plt.subplots(2)
ax1.hist(woa.c_t.where(woa.c_t).values.ravel(), bins=100, density=True, range=(-2,30),
weights=vol_full.where(woa.c_t).values.ravel());
ax2.hist(woa.s_a.where(woa.c_t).values.ravel(), bins=100, density=True, range=(32,38),
weights=vol_full.where(woa.s_a).values.ravel());
ax1.set_xlabel(r'Temperature ($^\circ$C)')
ax2.set_xlabel("Salinity (g / kg)")
plt.tight_layout()
plt.close()
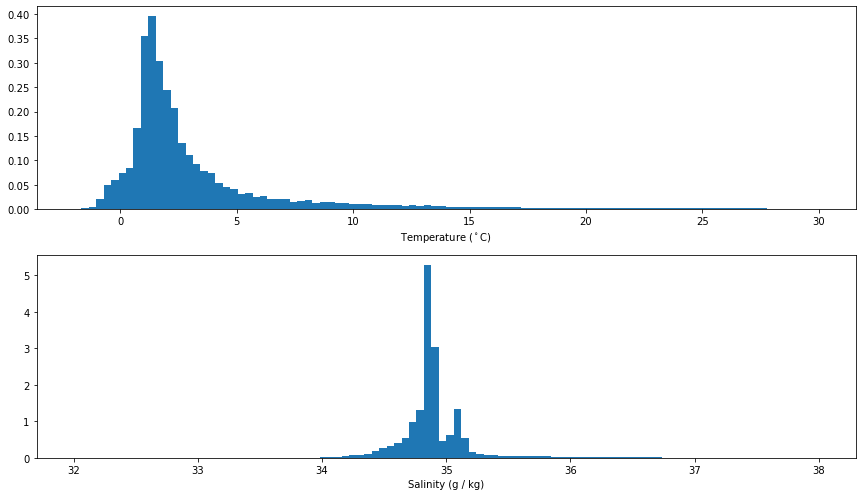
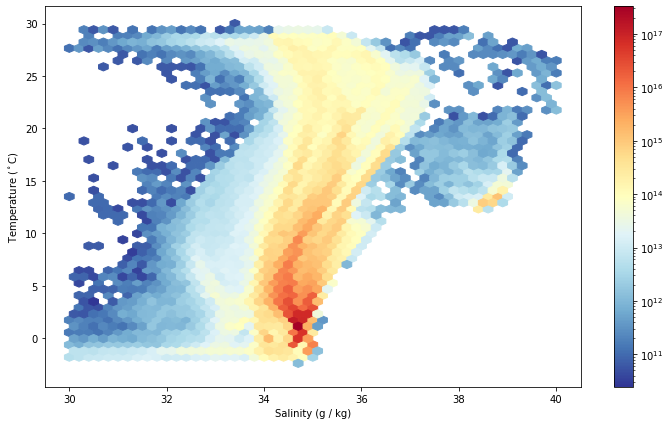
Temperature and Salinity Probability Distributions¶
fig
from matplotlib.colors import LogNorm
fig, ax = plt.subplots()
hb = ax.hexbin(woa.s_an.values.ravel(), woa.c_t.values.ravel(),
C=vol_full.values.ravel(), reduce_C_function=np.sum,
extent=(30,40,-3,30), gridsize=50, bins='log',
cmap='RdYlBu_r')
plt.colorbar(hb)
ax.set_ylabel(r'Temperature ($^\circ$C)')
ax.set_xlabel("Salinity (g / kg)")
plt.close()
Joint T/S Distribution¶
fig
#pal = seaborn.palettes.color_palette(n_colors=len(basins_main))
fig, ax = plt.subplots()
for (bnum, bname) in basins_main.items():
ax.plot(
woa.s_an.where(woa.basin==bnum).values.ravel(),
woa.c_t.where(woa.basin==bnum).values.ravel(),
'.', alpha=0.02, label=bname
)
ax.set_xlim(30,38)
ax.legend(loc='upper left', frameon=True)
plt.close()
# blue green red purple
T/S Distribution by Basin¶
fig
Thermohaline Circulation¶

# the goods are here
# https://www.nodc.noaa.gov/OC5/3M_HEAT_CONTENT/
# http://data.nodc.noaa.gov/thredds/catalog/woa/heat_content/heat_content/catalog.html
url = ('https://data.nodc.noaa.gov/thredds/dodsC/woa/heat_content/'
'heat_content/heat_content_anomaly_0-700_pentad.nc')
hc = xr.open_dataset(url, decode_times=False).load()
hc.time.attrs['calendar'] = '360_day'
hc = xr.decode_cf(hc)
hc
<xarray.Dataset>
Dimensions: (depth: 1, lat: 180, lon: 360, nbounds: 2, time: 62)
Coordinates:
* lat (lat) float32 -89.5 -88.5 -87.5 -86.5 ... 87.5 88.5 89.5
* lon (lon) float32 -179.5 -178.5 -177.5 ... 177.5 178.5 179.5
* time (time) object 1957-07-01 00:00:00 ... 2018-07-01 00:0...
Dimensions without coordinates: depth, nbounds
Data variables: (12/31)
crs int32 ...
lat_bnds (lat, nbounds) float32 ...
lon_bnds (lon, nbounds) float32 ...
depth_bnds (depth, nbounds) float32 ...
climatology_bounds (time, nbounds) float32 ...
h18_hc (time, depth, lat, lon) float32 ...
... ...
pent_h22_se_IO (time) float32 ...
pent_h22_NI (time) float32 ...
pent_h22_se_NI (time) float32 ...
pent_h22_SI (time) float32 ...
pent_h22_se_SI (time) float32 ...
basin_mask (lat, lon) float64 ...
Attributes: (12/45)
Conventions: CF-1.6
title: Ocean Heat Content anomalies from WOA09 ...
summary: Mean ocean variable anomaly from in situ...
references: Levitus, S., J. I. Antonov, T. P. Boyer,...
institution: National Oceanographic Data Center(NODC)
comment:
... ...
publisher_name: US NATIONAL OCEANOGRAPHIC DATA CENTER
publisher_url: http://www.nodc.noaa.gov/
publisher_email: NODC.Services@noaa.gov
license: These data are openly available to the p...
Metadata_Conventions: Unidata Dataset Discovery v1.0
metadata_link: http://www.nodc.noaa.gov/OC5/3M_HEAT_CON...- depth: 1
- lat: 180
- lon: 360
- nbounds: 2
- time: 62
- lat(lat)float32-89.5 -88.5 -87.5 ... 88.5 89.5
- standard_name :
- latitude
- long_name :
- latitude
- units :
- degrees_north
- axis :
- Y
- bounds :
- lat_bnds
array([-89.5, -88.5, -87.5, -86.5, -85.5, -84.5, -83.5, -82.5, -81.5, -80.5, -79.5, -78.5, -77.5, -76.5, -75.5, -74.5, -73.5, -72.5, -71.5, -70.5, -69.5, -68.5, -67.5, -66.5, -65.5, -64.5, -63.5, -62.5, -61.5, -60.5, -59.5, -58.5, -57.5, -56.5, -55.5, -54.5, -53.5, -52.5, -51.5, -50.5, -49.5, -48.5, -47.5, -46.5, -45.5, -44.5, -43.5, -42.5, -41.5, -40.5, -39.5, -38.5, -37.5, -36.5, -35.5, -34.5, -33.5, -32.5, -31.5, -30.5, -29.5, -28.5, -27.5, -26.5, -25.5, -24.5, -23.5, -22.5, -21.5, -20.5, -19.5, -18.5, -17.5, -16.5, -15.5, -14.5, -13.5, -12.5, -11.5, -10.5, -9.5, -8.5, -7.5, -6.5, -5.5, -4.5, -3.5, -2.5, -1.5, -0.5, 0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, 10.5, 11.5, 12.5, 13.5, 14.5, 15.5, 16.5, 17.5, 18.5, 19.5, 20.5, 21.5, 22.5, 23.5, 24.5, 25.5, 26.5, 27.5, 28.5, 29.5, 30.5, 31.5, 32.5, 33.5, 34.5, 35.5, 36.5, 37.5, 38.5, 39.5, 40.5, 41.5, 42.5, 43.5, 44.5, 45.5, 46.5, 47.5, 48.5, 49.5, 50.5, 51.5, 52.5, 53.5, 54.5, 55.5, 56.5, 57.5, 58.5, 59.5, 60.5, 61.5, 62.5, 63.5, 64.5, 65.5, 66.5, 67.5, 68.5, 69.5, 70.5, 71.5, 72.5, 73.5, 74.5, 75.5, 76.5, 77.5, 78.5, 79.5, 80.5, 81.5, 82.5, 83.5, 84.5, 85.5, 86.5, 87.5, 88.5, 89.5], dtype=float32) - lon(lon)float32-179.5 -178.5 ... 178.5 179.5
- standard_name :
- longitude
- long_name :
- longitude
- units :
- degrees_east
- axis :
- X
- bounds :
- lon_bnds
array([-179.5, -178.5, -177.5, ..., 177.5, 178.5, 179.5], dtype=float32)
- time(time)object1957-07-01 00:00:00 ... 2018-07-...
- standard_name :
- time
- long_name :
- time
- axis :
- T
- climatology :
- climatology_bounds
array([cftime.Datetime360Day(1957, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1958, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1959, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1960, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1961, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1962, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1963, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1964, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1965, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1966, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1967, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1968, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1969, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1970, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1971, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1972, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1973, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1974, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1975, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1976, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1977, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1978, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1979, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1980, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1981, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1982, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1983, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1984, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1985, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1986, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1987, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1988, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1989, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1990, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1991, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1992, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1993, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1994, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1995, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1996, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1997, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1998, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(1999, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2000, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2001, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2002, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2003, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2004, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2005, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2006, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2007, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2008, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2009, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2010, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2011, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2012, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2013, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2014, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2015, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2016, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2017, 7, 1, 0, 0, 0, 0, has_year_zero=False), cftime.Datetime360Day(2018, 7, 1, 0, 0, 0, 0, has_year_zero=False)], dtype=object)
- crs()int32...
- grid_mapping_name :
- latitude_longitude
- epsg_code :
- EPSG:4326
- longitude_of_prime_meridian :
- 0.0
- semi_major_axis :
- 6378137.0
- inverse_flattening :
- 298.25723
array(-2147483647, dtype=int32)
- lat_bnds(lat, nbounds)float32...
- comment :
- latitude bounds
array([[-90., -89.], [-89., -88.], [-88., -87.], ..., [ 87., 88.], [ 88., 89.], [ 89., 90.]], dtype=float32) - lon_bnds(lon, nbounds)float32...
- comment :
- longitude bounds
array([[-180., -179.], [-179., -178.], [-178., -177.], ..., [ 177., 178.], [ 178., 179.], [ 179., 180.]], dtype=float32) - depth_bnds(depth, nbounds)float32...
- comment :
- depth bounds
- positive :
- down
- units :
- meters
- axis :
- Z
array([[ 0., 700.]], dtype=float32)
- climatology_bounds(time, nbounds)float32...
- comment :
- This variable defines the bounds of the climatological time period for each time
array([[ 0., 60.], [ 12., 72.], [ 24., 84.], [ 36., 96.], [ 48., 108.], [ 60., 120.], [ 72., 132.], [ 84., 144.], [ 96., 156.], [108., 168.], [120., 180.], [132., 192.], [144., 204.], [156., 216.], [168., 228.], [180., 240.], [192., 252.], [204., 264.], [216., 276.], [228., 288.], [240., 300.], [252., 312.], [264., 324.], [276., 336.], [288., 348.], [300., 360.], [312., 372.], [324., 384.], [336., 396.], [348., 408.], [360., 420.], [372., 432.], [384., 444.], [396., 456.], [408., 468.], [420., 480.], [432., 492.], [444., 504.], [456., 516.], [468., 528.], [480., 540.], [492., 552.], [504., 564.], [516., 576.], [528., 588.], [540., 600.], [552., 612.], [564., 624.], [576., 636.], [588., 648.], [600., 660.], [612., 672.], [624., 684.], [636., 696.], [648., 708.], [660., 720.], [672., 732.], [684., 744.], [696., 756.], [708., 768.], [720., 780.], [732., 792.]], dtype=float32) - h18_hc(time, depth, lat, lon)float32...
- long_name :
- Ocean heat content anomaly calculated from objectively analyzed temperature anomaly fields over given depth limits
- coordinates :
- time lat lon depth
- cell_methods :
- area: mean depth: mean time: mean
- grid_mapping :
- crs
- units :
- 10^18_joules
[4017600 values with dtype=float32]
- pent_h22_WO(time)float32...
- long_name :
- global_heat_content_integral_0-700
- units :
- 10^22_joules
array([-6.288695, -4.892758, -5.784055, -3.387432, -2.707605, -2.603742, -3.997727, -4.665711, -5.374453, -7.158821, -7.297251, -6.883656, -5.412853, -4.618413, -3.601387, -3.221654, -3.128767, -2.439007, -1.457077, -0.977681, -0.182801, 0.975104, 1.579883, 1.113957, 0.523011, 0.486439, 0.21926 , 0.489551, 0.648078, 1.065748, 0.98001 , 0.760345, 0.803394, 1.565564, 2.711525, 3.7019 , 3.875865, 3.217625, 3.779482, 4.174185, 4.251321, 4.513308, 4.558651, 5.628954, 6.182415, 7.172529, 8.0118 , 9.127041, 9.104564, 9.314054, 9.451116, 10.22683 , 10.44346 , 10.641618, 11.493894, 11.878058, 12.916622, 13.396392, 14.142682, 14.800014, 14.903126, 15.740791], dtype=float32) - pent_h22_se_WO(time)float32...
- long_name :
- global_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([1.842686, 1.836877, 1.83623 , 1.71795 , 1.580727, 1.504552, 1.540631, 1.436218, 1.388474, 1.290961, 1.233682, 1.08622 , 0.979452, 0.913872, 0.88283 , 0.864919, 0.867636, 0.884624, 0.887177, 0.888699, 0.820958, 0.761577, 0.767867, 0.738683, 0.737726, 0.679426, 0.679605, 0.595538, 0.525312, 0.511351, 0.484396, 0.491693, 0.482232, 0.46751 , 0.39178 , 0.351994, 0.289757, 0.276638, 0.274458, 0.29157 , 0.303721, 0.348922, 0.409389, 0.428075, 0.367168, 0.282452, 0.217646, 0.194968, 0.179644, 0.171795, 0.168922, 0.161485, 0.162099, 0.157412, 0.141689, 0.142085, 0.136894, 0.134667, 0.131762, 0.13035 , 0.141381, 0.135071], dtype=float32) - pent_h22_NH(time)float32...
- long_name :
- northern_hemisphere_heat_content_integral_0-700
- units :
- 10^22_joules
array([-1.437559, -0.659048, -1.003371, -0.452752, -0.748635, -0.738145, -0.756852, -0.930149, -1.441751, -1.797886, -2.661819, -2.777363, -2.776239, -2.969397, -2.646983, -2.36598 , -1.957831, -1.877374, -1.399738, -1.264643, -0.59713 , -0.059998, 0.541609, 0.44218 , 0.391951, 0.716205, 0.471983, 0.315095, 0.374408, 0.488119, 0.28776 , 0.018627, 0.236868, 0.623496, 0.874215, 1.145203, 1.656924, 1.980619, 2.003117, 2.59605 , 2.880694, 3.27235 , 3.466433, 3.871615, 4.120043, 4.569418, 4.950961, 5.444228, 5.637978, 5.783906, 5.671659, 5.729272, 5.59363 , 5.591918, 5.759031, 5.641775, 5.876155, 6.14617 , 6.522678, 6.723238, 6.562952, 7.659342], dtype=float32) - pent_h22_se_NH(time)float32...
- long_name :
- northern_hemisphere_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([0.760144, 0.708205, 0.669029, 0.612442, 0.568487, 0.49362 , 0.512772, 0.423674, 0.384655, 0.348872, 0.313734, 0.23007 , 0.187796, 0.157003, 0.148355, 0.15313 , 0.147873, 0.160858, 0.16389 , 0.160115, 0.1538 , 0.127889, 0.128299, 0.125208, 0.123709, 0.122135, 0.120513, 0.102694, 0.096655, 0.086172, 0.078795, 0.082491, 0.075917, 0.074797, 0.070494, 0.069148, 0.07437 , 0.080821, 0.085599, 0.094038, 0.097038, 0.100984, 0.10323 , 0.097369, 0.089892, 0.085449, 0.082612, 0.087238, 0.084533, 0.083093, 0.082276, 0.076453, 0.0781 , 0.077609, 0.065256, 0.06688 , 0.064154, 0.061274, 0.059592, 0.059999, 0.071174, 0.065938], dtype=float32) - pent_h22_SH(time)float32...
- long_name :
- southern_hemisphere_heat_content_integral_0-700
- units :
- 10^22_joules
array([-4.851153, -4.233699, -4.780672, -2.934695, -1.958968, -1.865586, -3.240879, -3.735567, -3.932683, -5.36095 , -4.635412, -4.106285, -2.636613, -1.649003, -0.954393, -0.855672, -1.170939, -0.561638, -0.057339, 0.28696 , 0.414331, 1.035097, 1.038276, 0.671779, 0.131058, -0.229767, -0.252721, 0.174455, 0.27367 , 0.577621, 0.692252, 0.741719, 0.566525, 0.942066, 1.837317, 2.556699, 2.21895 , 1.23701 , 1.776369, 1.578133, 1.370625, 1.240958, 1.092215, 1.757334, 2.062383, 2.603179, 3.060834, 3.682796, 3.466569, 3.530176, 3.779429, 4.497595, 4.849782, 5.049636, 5.734864, 6.236268, 7.040561, 7.250291, 7.62019 , 8.076709, 8.340103, 8.081578], dtype=float32) - pent_h22_se_SH(time)float32...
- long_name :
- southern_hemisphere_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([1.082542, 1.128671, 1.167202, 1.105508, 1.01224 , 1.010932, 1.02786 , 1.012545, 1.003818, 0.942089, 0.919947, 0.85615 , 0.791656, 0.756868, 0.734475, 0.711789, 0.719763, 0.723766, 0.723287, 0.728584, 0.667159, 0.633688, 0.639568, 0.613475, 0.614017, 0.557291, 0.559092, 0.492845, 0.428657, 0.425179, 0.405601, 0.409202, 0.406315, 0.392713, 0.321286, 0.282847, 0.215387, 0.195817, 0.188859, 0.197532, 0.206683, 0.247938, 0.306159, 0.330706, 0.277276, 0.197002, 0.135034, 0.107729, 0.095111, 0.088702, 0.086646, 0.085032, 0.083999, 0.079802, 0.076433, 0.075205, 0.072741, 0.073392, 0.072169, 0.070351, 0.070207, 0.069133], dtype=float32) - pent_h22_AO(time)float32...
- long_name :
- Atlantic_Ocean_heat_content_integral_0-700
- units :
- 10^22_joules
array([-2.601018, -2.403772, -2.550205, -1.655523, -2.049075, -1.376147, -2.029487, -1.769845, -2.0978 , -2.047394, -2.409131, -2.255042, -1.917121, -1.611235, -1.634703, -1.697296, -1.629409, -1.333255, -1.122638, -1.389839, -0.871172, -0.530094, -0.672035, -0.38288 , 0.086118, 0.198783, 0.097296, 0.211876, 0.196669, 0.013917, 0.049612, -0.081866, 0.224367, 0.5009 , 0.902423, 1.191282, 1.685269, 1.872183, 1.951509, 2.084828, 2.13143 , 1.610081, 1.72136 , 2.449398, 2.798403, 3.09352 , 3.968912, 4.480746, 4.679378, 4.646803, 4.564225, 4.71802 , 4.515999, 4.504152, 4.792003, 5.031074, 5.243983, 5.471953, 5.701963, 6.067047, 5.749677, 6.622941], dtype=float32) - pent_h22_se_AO(time)float32...
- long_name :
- Atlantic_Ocean_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([0.549281, 0.451574, 0.459504, 0.425795, 0.384527, 0.357041, 0.397102, 0.411782, 0.413791, 0.391557, 0.360218, 0.293461, 0.268265, 0.233445, 0.215042, 0.228016, 0.229463, 0.22407 , 0.233156, 0.22162 , 0.171912, 0.15807 , 0.158374, 0.154039, 0.139462, 0.1364 , 0.121027, 0.108742, 0.104005, 0.11295 , 0.10241 , 0.08628 , 0.078914, 0.071998, 0.062929, 0.062805, 0.060988, 0.062515, 0.066541, 0.077938, 0.080489, 0.093129, 0.117507, 0.117427, 0.085546, 0.079309, 0.065084, 0.065984, 0.06624 , 0.064463, 0.063254, 0.060165, 0.061565, 0.058764, 0.047997, 0.047022, 0.043588, 0.042949, 0.042403, 0.041891, 0.052399, 0.044536], dtype=float32) - pent_h22_NA(time)float32...
- long_name :
- North_Atlantic_heat_content_integral_0-700
- units :
- 10^22_joules
array([-0.971802, -0.658995, -0.669964, -0.344899, -0.659023, -0.623111, -0.820188, -0.820973, -0.985068, -0.992146, -1.316494, -1.385485, -1.291805, -1.161226, -1.156855, -1.079825, -1.017811, -0.96968 , -0.865807, -0.756418, -0.431954, -0.161428, -0.181038, -0.154193, -0.09906 , -0.050861, -0.20386 , -0.144289, -0.11867 , -0.04111 , -0.092286, -0.096791, 0.114696, 0.317155, 0.481932, 0.643853, 0.93214 , 1.096446, 1.182772, 1.513879, 1.650899, 1.526848, 1.697952, 2.081093, 2.229178, 2.528198, 2.926768, 3.246055, 3.425094, 3.364953, 3.21241 , 3.189281, 3.065194, 3.076496, 3.039903, 3.122022, 3.185949, 3.237285, 3.408746, 3.548848, 3.089961, 3.874815], dtype=float32) - pent_h22_se_NA(time)float32...
- long_name :
- North_Atlantic_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([0.323197, 0.22556 , 0.207545, 0.171924, 0.168304, 0.152904, 0.170345, 0.164224, 0.166242, 0.14454 , 0.127696, 0.084085, 0.067969, 0.053195, 0.047979, 0.041452, 0.045811, 0.047482, 0.048702, 0.047387, 0.049432, 0.047692, 0.046142, 0.044978, 0.043176, 0.041504, 0.036092, 0.036152, 0.034219, 0.031592, 0.029042, 0.03352 , 0.031477, 0.032242, 0.028285, 0.026588, 0.028339, 0.029799, 0.027763, 0.027874, 0.029225, 0.029812, 0.029883, 0.030857, 0.029618, 0.029933, 0.030248, 0.03963 , 0.042702, 0.041764, 0.04234 , 0.03864 , 0.039652, 0.038327, 0.028578, 0.028307, 0.026971, 0.024828, 0.024125, 0.023592, 0.03384 , 0.027145], dtype=float32) - pent_h22_SA(time)float32...
- long_name :
- South_Atlantic_heat_content_integral_0-700
- units :
- 10^22_joules
array([-1.62921 , -1.744777, -1.880237, -1.310617, -1.390049, -0.753037, -1.209292, -0.948869, -1.112738, -1.055247, -1.092641, -0.869553, -0.625331, -0.450013, -0.477848, -0.617477, -0.611598, -0.363576, -0.256833, -0.63342 , -0.439217, -0.368667, -0.490996, -0.228687, 0.185177, 0.249645, 0.301157, 0.356167, 0.315339, 0.055027, 0.141898, 0.014926, 0.10967 , 0.183746, 0.420488, 0.547429, 0.753134, 0.775732, 0.768739, 0.570943, 0.480527, 0.083232, 0.023408, 0.368303, 0.569223, 0.565323, 1.042145, 1.234701, 1.254277, 1.281852, 1.351795, 1.528761, 1.450815, 1.427677, 1.752088, 1.909044, 2.058082, 2.234727, 2.293216, 2.518219, 2.659722, 2.748131], dtype=float32) - pent_h22_se_SA(time)float32...
- long_name :
- South_Atlantic_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([0.226085, 0.226015, 0.251959, 0.253871, 0.216223, 0.204137, 0.226757, 0.247558, 0.247549, 0.247018, 0.232522, 0.209376, 0.200296, 0.18025 , 0.167063, 0.186564, 0.183651, 0.176589, 0.184454, 0.174232, 0.12248 , 0.110379, 0.112232, 0.109061, 0.096286, 0.094896, 0.084935, 0.072589, 0.069786, 0.081358, 0.073368, 0.052761, 0.047437, 0.039756, 0.034644, 0.036218, 0.032649, 0.032717, 0.038778, 0.050064, 0.051264, 0.063317, 0.087624, 0.08657 , 0.055929, 0.049376, 0.034836, 0.026354, 0.023538, 0.022699, 0.020914, 0.021525, 0.021912, 0.020436, 0.019419, 0.018715, 0.016616, 0.018121, 0.018279, 0.018299, 0.018559, 0.017391], dtype=float32) - pent_h22_PO(time)float32...
- long_name :
- Pacific_Ocean_heat_content_integral_0-700
- units :
- 10^22_joules
array([-2.505641, -1.258843, -1.825778, -0.671822, 0.046691, -0.422522, -0.687086, -1.6234 , -1.964979, -2.7014 , -2.182975, -2.388641, -1.671371, -1.539451, -1.42994 , -1.072202, -1.031902, -0.921352, -0.468627, 0.33969 , -0.049241, 0.65199 , 1.213085, 0.892058, -0.087446, -0.114917, -0.421509, -0.406992, -0.479219, -0.01708 , -0.20132 , -0.420039, -0.61305 , 0.167696, 0.811469, 1.266996, 1.483416, 1.401909, 1.495817, 1.73941 , 1.799246, 2.356171, 2.214761, 2.234168, 2.432663, 3.03216 , 3.191943, 3.784683, 3.33058 , 3.397692, 3.30255 , 3.371164, 3.253877, 3.332361, 3.513103, 3.487733, 4.076708, 4.296066, 4.884754, 5.352047, 5.774701, 6.011369], dtype=float32) - pent_h22_se_PO(time)float32...
- long_name :
- Pacific_Ocean_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([0.852305, 0.907993, 0.905732, 0.877106, 0.81095 , 0.798834, 0.787108, 0.685402, 0.596136, 0.510756, 0.484833, 0.40036 , 0.36065 , 0.345753, 0.347684, 0.341102, 0.375639, 0.395838, 0.423107, 0.437426, 0.386734, 0.356969, 0.360383, 0.338509, 0.355802, 0.320181, 0.336998, 0.291861, 0.250878, 0.233666, 0.206732, 0.207847, 0.207638, 0.169675, 0.151747, 0.147551, 0.137243, 0.127858, 0.134129, 0.147953, 0.159924, 0.181806, 0.203311, 0.214662, 0.197079, 0.146415, 0.110158, 0.094441, 0.082387, 0.078366, 0.077428, 0.073563, 0.072285, 0.069551, 0.066323, 0.067268, 0.065255, 0.063972, 0.061715, 0.061801, 0.062588, 0.063597], dtype=float32) - pent_h22_NP(time)float32...
- long_name :
- North_Pacific_heat_content_integral_0-700
- units :
- 10^22_joules
array([-0.396412, 0.065448, -0.130504, 0.0774 , -0.046284, -0.094239, 0.149377, -0.026569, -0.346036, -0.622329, -1.12759 , -1.22611 , -1.366167, -1.75808 , -1.458844, -1.226601, -0.934491, -0.900025, -0.508825, -0.529617, -0.193507, 0.057029, 0.612717, 0.474969, 0.355347, 0.608038, 0.423245, 0.338217, 0.267873, 0.392955, 0.208055, -0.100985, -0.117688, 0.054884, 0.081157, 0.203569, 0.485357, 0.697317, 0.691699, 1.02005 , 1.250703, 1.856393, 1.946206, 1.912065, 1.935059, 1.932052, 1.887404, 2.051608, 2.062479, 2.227767, 2.225039, 2.225552, 2.133141, 2.026886, 2.138745, 1.905073, 2.0191 , 2.134571, 2.366668, 2.48133 , 2.746387, 3.046807], dtype=float32) - pent_h22_se_NP(time)float32...
- long_name :
- North_Pacific_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([0.363998, 0.411451, 0.408016, 0.397239, 0.374058, 0.322116, 0.324826, 0.241054, 0.199488, 0.179092, 0.16322 , 0.123118, 0.100378, 0.084312, 0.084435, 0.088233, 0.085304, 0.089655, 0.101005, 0.102614, 0.097126, 0.073674, 0.075916, 0.073281, 0.072559, 0.067909, 0.07267 , 0.05613 , 0.050528, 0.046164, 0.041731, 0.041099, 0.036784, 0.035429, 0.035555, 0.036889, 0.040901, 0.046116, 0.052928, 0.060996, 0.062301, 0.064264, 0.065076, 0.057896, 0.053229, 0.049947, 0.047404, 0.042927, 0.037312, 0.037048, 0.035337, 0.033109, 0.033543, 0.033646, 0.031936, 0.03338 , 0.031868, 0.031127, 0.03018 , 0.031965, 0.032916, 0.033831], dtype=float32) - pent_h22_SP(time)float32...
- long_name :
- South_Pacific_heat_content_integral_0-700
- units :
- 10^22_joules
array([-2.109229, -1.324292, -1.695278, -0.749222, 0.092974, -0.328284, -0.836464, -1.596832, -1.61895 , -2.079066, -1.055388, -1.162532, -0.305203, 0.218624, 0.028904, 0.154399, -0.097411, -0.021326, 0.040198, 0.869309, 0.144267, 0.594961, 0.600366, 0.417086, -0.442792, -0.722954, -0.844751, -0.745209, -0.747093, -0.410037, -0.409375, -0.319054, -0.495362, 0.112811, 0.730311, 1.063425, 0.99806 , 0.704595, 0.804119, 0.719358, 0.548543, 0.499778, 0.268558, 0.322102, 0.497605, 1.100108, 1.304535, 1.73308 , 1.268098, 1.16992 , 1.077507, 1.145603, 1.120738, 1.305473, 1.374368, 1.582664, 2.057606, 2.161493, 2.518093, 2.870713, 3.028324, 2.964572], dtype=float32) - pent_h22_se_SP(time)float32...
- long_name :
- South_Pacific_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([0.488307, 0.496542, 0.497716, 0.479867, 0.436892, 0.476718, 0.462282, 0.444348, 0.396648, 0.331664, 0.321613, 0.277242, 0.260271, 0.261441, 0.263249, 0.252869, 0.290336, 0.306183, 0.322102, 0.334812, 0.289608, 0.283295, 0.284467, 0.265228, 0.283243, 0.252271, 0.264328, 0.235731, 0.20035 , 0.187502, 0.165001, 0.166749, 0.170854, 0.134246, 0.116191, 0.110662, 0.096342, 0.081742, 0.081201, 0.086957, 0.097622, 0.117542, 0.138235, 0.156767, 0.143851, 0.096468, 0.062754, 0.051514, 0.045075, 0.041319, 0.042091, 0.040454, 0.038742, 0.035905, 0.034387, 0.033888, 0.033387, 0.032845, 0.031536, 0.029836, 0.029673, 0.029765], dtype=float32) - pent_h22_IO(time)float32...
- long_name :
- Indian_Ocean_heat_content_integral_0-700
- units :
- 10^22_joules
array([-1.190544, -1.237827, -1.415187, -1.067977, -0.714793, -0.814509, -1.28986 , -1.281453, -1.320256, -2.415456, -2.709558, -2.239426, -1.822071, -1.454605, -0.530236, -0.447161, -0.46054 , -0.177183, 0.13932 , 0.077665, 0.741772, 0.856154, 1.039237, 0.604857, 0.52622 , 0.406276, 0.548503, 0.692143, 0.935871, 1.071709, 1.133769, 1.263005, 1.191998, 0.89961 , 1.002419, 1.244923, 0.707896, -0.055429, 0.333062, 0.349482, 0.321797, 0.549685, 0.624599, 0.939241, 0.946499, 1.043774, 0.848112, 0.859415, 1.092657, 1.26719 , 1.580536, 2.132398, 2.671292, 2.800793, 3.176493, 3.34448 , 3.581322, 3.615808, 3.542609, 3.372986, 3.366693, 3.094792], dtype=float32) - pent_h22_se_IO(time)float32...
- long_name :
- Indian_Ocean_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([0.440468, 0.476714, 0.470514, 0.414549, 0.384633, 0.348174, 0.355838, 0.338394, 0.377956, 0.388035, 0.387996, 0.391965, 0.350018, 0.333657, 0.319396, 0.295149, 0.261901, 0.264159, 0.230418, 0.229163, 0.261832, 0.246197, 0.248726, 0.245772, 0.242061, 0.222393, 0.221112, 0.194414, 0.169899, 0.164385, 0.174892, 0.197212, 0.195308, 0.225493, 0.176769, 0.141355, 0.091226, 0.085974, 0.07349 , 0.065355, 0.062921, 0.073617, 0.088133, 0.09544 , 0.084013, 0.056319, 0.042022, 0.034138, 0.030612, 0.028557, 0.027731, 0.027221, 0.027526, 0.027587, 0.026607, 0.026597, 0.026655, 0.026263, 0.026152, 0.026091, 0.025837, 0.02637 ], dtype=float32) - pent_h22_NI(time)float32...
- long_name :
- North_Indian_heat_content_integral_0-700
- units :
- 10^22_joules
array([-7.78040e-02, -7.31980e-02, -2.10055e-01, -1.93125e-01, -5.28990e-02, -3.02480e-02, -9.47420e-02, -9.15920e-02, -1.19249e-01, -1.88818e-01, -2.22175e-01, -1.65207e-01, -1.15987e-01, -3.69970e-02, -2.47900e-02, -5.45700e-02, 1.38900e-03, -4.46000e-04, -1.99760e-02, 2.65910e-02, 3.24930e-02, 4.73550e-02, 1.10331e-01, 1.21477e-01, 1.37544e-01, 1.62733e-01, 2.57633e-01, 1.28646e-01, 2.30449e-01, 1.39076e-01, 1.74040e-01, 2.17159e-01, 2.39781e-01, 2.54098e-01, 3.15908e-01, 2.99078e-01, 2.40138e-01, 1.87890e-01, 1.29556e-01, 6.16390e-02, -1.97630e-02, -1.08264e-01, -1.75640e-01, -1.27694e-01, -4.90640e-02, 1.06031e-01, 1.33955e-01, 1.44411e-01, 1.48459e-01, 1.88783e-01, 2.30404e-01, 3.09154e-01, 3.93063e-01, 4.84315e-01, 5.68080e-01, 5.99922e-01, 6.56450e-01, 7.61710e-01, 7.33754e-01, 6.85185e-01, 7.14631e-01, 7.25891e-01], dtype=float32) - pent_h22_se_NI(time)float32...
- long_name :
- North_Indian_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([0.072317, 0.070599, 0.052988, 0.04278 , 0.025508, 0.018097, 0.017017, 0.017756, 0.018334, 0.024627, 0.022184, 0.022433, 0.018928, 0.018479, 0.015233, 0.022793, 0.016125, 0.023165, 0.013687, 0.009623, 0.006762, 0.006182, 0.005856, 0.006587, 0.007574, 0.01227 , 0.011282, 0.009889, 0.011379, 0.008066, 0.00766 , 0.007519, 0.007284, 0.006783, 0.006319, 0.005387, 0.00483 , 0.004615, 0.004609, 0.004844, 0.005124, 0.006538, 0.007832, 0.008071, 0.006516, 0.00516 , 0.004578, 0.004276, 0.004114, 0.003873, 0.00409 , 0.004168, 0.004182, 0.004125, 0.003979, 0.003995, 0.003917, 0.003836, 0.003797, 0.003874, 0.003862, 0.004394], dtype=float32) - pent_h22_SI(time)float32...
- long_name :
- South_Indian_heat_content_integral_0-700
- units :
- 10^22_joules
array([-1.112741, -1.164628, -1.205132, -0.874852, -0.661894, -0.78426 , -1.195117, -1.18986 , -1.201008, -2.226636, -2.487383, -2.07422 , -1.706083, -1.417609, -0.505447, -0.392591, -0.46193 , -0.176736, 0.159296, 0.051074, 0.709279, 0.8088 , 0.928907, 0.48338 , 0.388676, 0.243543, 0.29087 , 0.563496, 0.705422, 0.932633, 0.959728, 1.045848, 0.952217, 0.645512, 0.686511, 0.945844, 0.467759, -0.24332 , 0.203506, 0.287843, 0.34156 , 0.657949, 0.80024 , 1.066936, 0.995562, 0.937744, 0.714157, 0.715005, 0.944198, 1.078406, 1.350131, 1.82324 , 2.278228, 2.31648 , 2.608411, 2.744557, 2.924873, 2.854093, 2.808856, 2.687801, 2.65206 , 2.368902], dtype=float32) - pent_h22_se_SI(time)float32...
- long_name :
- South_Indian_standarderror_heat_content_integral_0-700
- units :
- 10^22_joules
array([0.368151, 0.406115, 0.417526, 0.371769, 0.359125, 0.330077, 0.338821, 0.320639, 0.359621, 0.363408, 0.365812, 0.369532, 0.331089, 0.315178, 0.304163, 0.272355, 0.245776, 0.240995, 0.216731, 0.21954 , 0.25507 , 0.240015, 0.24287 , 0.239185, 0.234488, 0.210123, 0.209829, 0.184525, 0.158521, 0.156318, 0.167232, 0.189693, 0.188024, 0.21871 , 0.170451, 0.135967, 0.086396, 0.081359, 0.06888 , 0.060511, 0.057797, 0.067079, 0.0803 , 0.087369, 0.077497, 0.051158, 0.037444, 0.029861, 0.026498, 0.024684, 0.023641, 0.023053, 0.023344, 0.023461, 0.022627, 0.022602, 0.022738, 0.022427, 0.022355, 0.022217, 0.021975, 0.021977], dtype=float32) - basin_mask(lat, lon)float64...
- long_name :
- basin mask
- grid_mapping :
- crs
- flag_values :
- Not_used_in_Basin_Integrals Atlantic_Basin Pacific_Basin Indian_Basin
- comment :
- For North Hem. use Lats 0.5 to 89.5 For South use Lats -89.5 to -0.5
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.], [ 1., 1., 1., ..., 1., 1., 1.]])
- Conventions :
- CF-1.6
- title :
- Ocean Heat Content anomalies from WOA09 : heat_content_anomaly 0-700 m pentad 1.00 degree
- summary :
- Mean ocean variable anomaly from in situ profile data
- references :
- Levitus, S., J. I. Antonov, T. P. Boyer, O. K. Baranova, H. E. Garcia, R. A. Locarnini, A.V. Mishonov, J. R. Reagan, D. Seidov, E. S. Yarosh, M. M. Zweng, 2012: World Ocean heat content and thermosteric sea level change (0-2000 m) 1955-2010. Geophys. Res. Lett. , 39, L10603, doi:10.1029/2012GL051106
- institution :
- National Oceanographic Data Center(NODC)
- comment :
- id :
- heat_content_anomaly_0-700_pentad.nc
- naming_authority :
- gov.noaa.nodc
- time_coverage_start :
- 1955-01-01
- time_coverage_duration :
- P66Y
- time_coverage_resolution :
- P05Y
- geospatial_lat_min :
- -90.0
- geospatial_lat_max :
- 90.0
- geospatial_lon_min :
- -180.0
- geospatial_lon_max :
- 540.0
- geospatial_vertical_min :
- 0.0
- geospatial_vertical_max :
- 700.0
- geospatial_lat_units :
- degrees_north
- geospatial_lat_resolution :
- 1.00 degrees
- geospatial_lon_units :
- degrees_east
- geospatial_lon_resolution :
- 1.00 degrees
- geospatial_vertical_units :
- m
- geospatial_vertical_resolution :
- geospatial_vertical_positive :
- down
- creator_name :
- Ocean Climate Laboratory
- creator_email :
- NODC.Services@noaa.gov
- creator_url :
- http://www.nodc.noaa.gov
- project :
- World Ocean Database
- processing_level :
- processed
- keywords :
- <ISO_TOPIC_Category> Oceans</ISO_TOPIC_Category>
- keywords_vocabulary :
- ISO 19115
- standard_name_vocabulary :
- CF-1.6
- contributor_name :
- Ocean Climate Laboratory
- contributor_role :
- Calculation of anomalies
- featureType :
- Grid
- cdm_data_type :
- Grid
- nodc_template_version :
- NODC_NetCDF_Grid_Template_v1.0
- date_created :
- 2021-01-11
- date_modified :
- 2021-01-11
- publisher_name :
- US NATIONAL OCEANOGRAPHIC DATA CENTER
- publisher_url :
- http://www.nodc.noaa.gov/
- publisher_email :
- NODC.Services@noaa.gov
- license :
- These data are openly available to the public. Please acknowledge the use of these data with the text given in the acknowledgment attribute.
- Metadata_Conventions :
- Unidata Dataset Discovery v1.0
- metadata_link :
- http://www.nodc.noaa.gov/OC5/3M_HEAT_CONTENT/
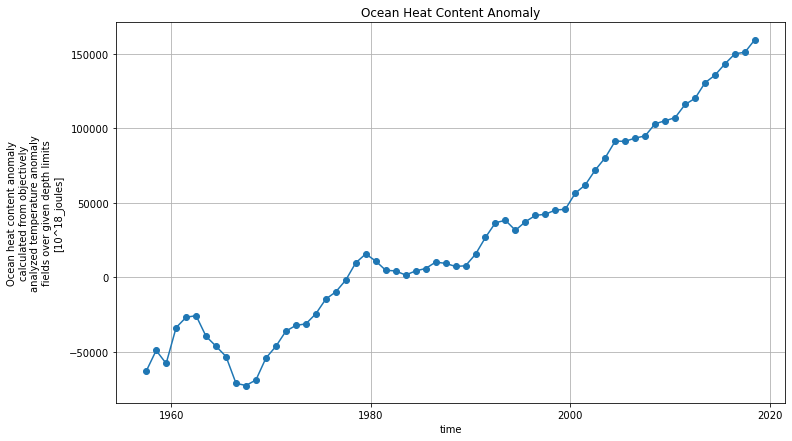
fig, ax = plt.subplots()
hc['h18_hc'].sum(dim=('lon','lat','depth'), keep_attrs=True).plot(ax=ax, marker='o')
plt.title('Ocean Heat Content Anomaly')
plt.grid()
plt.close()
Ocean Warming¶
We have been looking at climatologies. But we can’t forget that the ocean is warming!
Data from https://www.nodc.noaa.gov/OC5/3M_HEAT_CONTENT/.
fig