Numpy and Matplotlib¶
These are two of the most fundamental parts of the scientific python "ecosystem". Most everything else is built on top of them.
import numpy as np
What did we just do? We imported a package. This brings new variables (mostly functions) into our interpreter. We access them as follows.
# find out what is in our namespace
dir()
# find out what's in numpy
dir(np)
# find out what version we have
np.__version__
The numpy documentation is crucial!
http://docs.scipy.org/doc/numpy/reference/
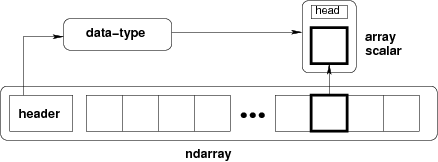
NDArrays¶
The core class is the numpy ndarray (n-dimensional array).
from IPython.display import Image
Image(url='http://docs.scipy.org/doc/numpy/_images/threefundamental.png')
# create an array from a list
a = np.array([9,0,2,1,0])
# find out the datatype
a.dtype
# find out the shape
a.shape
# what is the shape
type(a.shape)
# another array with a different datatype and shape
b = np.array([[5,3,1,9],[9,2,3,0]], dtype=np.float64)
# check dtype and shape
b.dtype, b.shape
Important Concept: The fastest varying dimension is the last dimension! The outer level of the hierarchy is the first dimension. (This is called "c-style" indexing)
More array creation¶
There are lots of ways to create arrays.
# create some uniform arrays
c = np.zeros((9,9))
d = np.ones((3,6,3), dtype=np.complex128)
e = np.full((3,3), np.pi)
e = np.ones_like(c)
f = np.zeros_like(d)
# create some ranges
np.arange(10)
# arange is left inclusive, right exclusive
np.arange(2,4,0.25)
# linearly spaced
np.linspace(2,4,20)
# log spaced
np.logspace(1,2,10)
# two dimensional grids
x = np.linspace(-2*np.pi, 2*np.pi, 100)
y = np.linspace(-np.pi, np.pi, 50)
xx, yy = np.meshgrid(x, y)
xx.shape, yy.shape
Indexing¶
Basic indexing is similar to lists
# get some individual elements of xx
xx[0,0], xx[-1,-1], xx[3,-5]
# get some whole rows and columns
xx[0].shape, xx[:,-1].shape
# get some ranges
xx[3:10,30:40].shape
There are many advanced ways to index arrays. You can read about them in the manual. Here is one example.
# use a boolean array as an index
idx = xx<0
yy[idx].shape
# the array got flattened
xx.ravel().shape
f = np.sin(xx) * np.cos(0.5*yy)
At this point you might be getting curious what these arrays "look" like. So we need to introduce some visualization.
from matplotlib import pyplot as plt
%matplotlib inline
plt.pcolormesh(f)
Manipulating array dimensions¶
# transpose
plt.pcolormesh(f.T)
# reshape an array (wrong size)
g = np.reshape(f, (8,9))
# reshape an array (right size) and mess it up
print(f.size)
g = np.reshape(f, (200,25))
plt.pcolormesh(g)
# tile an array
plt.pcolormesh(np.tile(f,(6,1)))
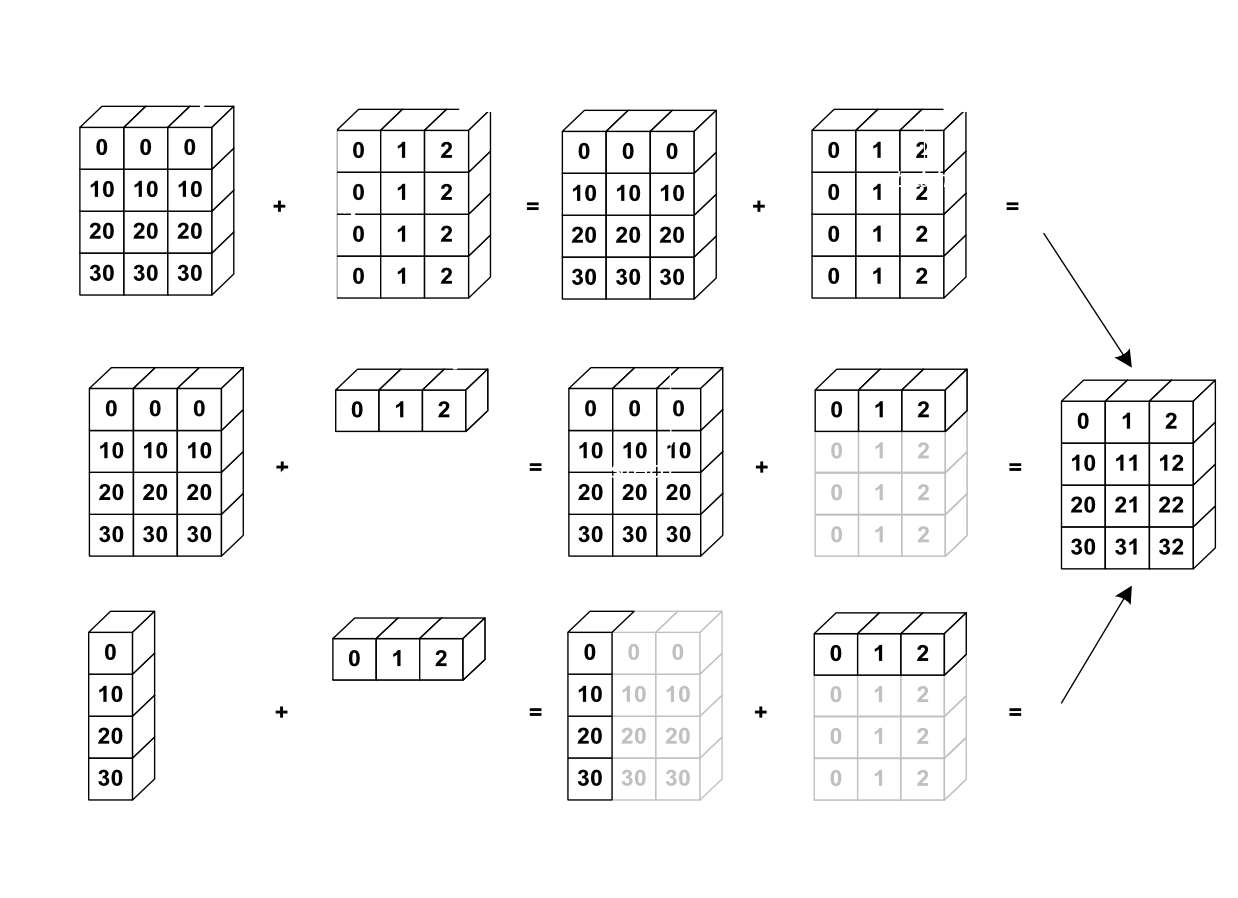
Broadcasting¶
Broadcasting is an efficient way to multiply arrays of different sizes
Image(url='http://scipy-lectures.github.io/_images/numpy_broadcasting.png',
width=720)
# multiply f by x
print(f.shape, x.shape)
g = f * x
print(g.shape)
# multiply f by y
print(f.shape, y.shape)
h = f * y
print(h.shape)
# use newaxis special syntax
h = f * y[:,np.newaxis]
print(h.shape)
plt.pcolormesh(g)
Reduction Operations¶
# sum
g.sum()
# mean
g.mean()
# std
g.std()
# apply on just one axis
g_ymean = g.mean(axis=0)
g_xmean = g.mean(axis=1)
plt.plot(x, g_ymean)
plt.plot(g_xmean, y)
Fancy Plotting¶
Enough lessons, let's have some fun.
fig = plt.figure(figsize=(12,8))
ax1 = plt.subplot2grid((6,6),(0,1),colspan=5)
ax2 = plt.subplot2grid((6,6),(1,0),rowspan=5)
fig = plt.figure(figsize=(10,6))
ax1 = plt.subplot2grid((6,6),(0,1),colspan=5)
ax2 = plt.subplot2grid((6,6),(1,0),rowspan=5)
ax3 = plt.subplot2grid((6,6),(1,1),rowspan=5, colspan=5)
ax1.plot(x, g_ymean)
ax2.plot(g_xmean, y)
ax3.pcolormesh(x, y, g)
ax1.set_xlim([x.min(), x.max()])
ax3.set_xlim([x.min(), x.max()])
ax2.set_ylim([y.min(), y.max()])
ax3.set_ylim([y.min(), y.max()])
plt.tight_layout()
Real Data¶
ARGO float profile from North Atlantic
# download with curl
!curl -O https://www.ldeo.columbia.edu/~rpa/argo_float_4901412.npz
# load numpy file and examine keys
data = np.load('argo_float_4901412.npz')
data.keys()
# access some data
T = data['T']
# there are "nans", missing data, which screw up our routines
T.min()
ar_w_mask = np.ma.masked_array([1, 2, 3, 4, 5],
mask=[True, True, False, False, False])
ar_w_mask
ar_w_mask.mean()
T_ma = np.ma.masked_invalid(T)
T_ma.mean()
Masked Arrays¶
This is how we deal with missing data in numpy
# create masked array
T = np.ma.masked_invalid(data['T'])
type(T)
# max and min
T.max(), T.min()
# load other data
S = np.ma.masked_invalid(data['S'])
P = np.ma.masked_invalid(data['P'])
# scatter plot
plt.scatter(S, T, c=P)
plt.grid()
plt.colorbar()