Xarray for multidimensional gridded data¶
In last week's lecture, we saw how Pandas provided a way to keep track of additional "metadata" surrounding tabular datasets, including "indexes" for each row and labels for each column. These features, together with Pandas' many useful routines for all kinds of data munging and analysis, have made Pandas one of the most popular python packages in the world.
However, not all Earth science datasets easily fit into the "tabular" model (i.e. rows and columns) imposed by Pandas. In particular, we often deal with multidimensional data. By multidimensional data (also often called N-dimensional), I mean data with many independent dimensions or axes. For example, we might represent Earth's surface temperature $T$ as a three dimensional variable
$$ T(x, y, t) $$
where $x$ is longitude, $y$ is latitude, and $t$ is time.
The point of xarray is to provide pandas-level convenience for working with this type of data.

NOTE: In order to run this tutorial, you need xarray and netCDF4 packages installed. The best thing to do is to create a custom conda environment, as described on the python installation page (scroll to Geosciences Python Environment). To test whether your environment is set up properly, try the following imports:
import xarray
import netCDF4
Xarray data structures¶
Like Pandas, xarray has two fundamental data structures:
- a
DataArray, which holds a single multi-dimensional variable and its coordinates - a
Dataset, which holds multiple variables that potentially share the same coordinates
DataArray¶
A DataArray has four essential attributes:
values: anumpy.ndarrayholding the array’s valuesdims: dimension names for each axis (e.g.,('x', 'y', 'z'))coords: a dict-like container of arrays (coordinates) that label each point (e.g., 1-dimensional arrays of numbers, datetime objects or strings)attrs: anOrderedDictto hold arbitrary metadata (attributes)
Let's start by constructing some DataArrays manually
import numpy as np
import xarray as xr
from matplotlib import pyplot as plt
%matplotlib inline
plt.rcParams['figure.figsize'] = (8,5)
A simple DataArray without dimensions or coordinates isn't much use.
da = xr.DataArray([9, 0, 2, 1, 0])
da
We can add a dimension name...
da = xr.DataArray([9, 0, 2, 1, 0], dims=['x'])
da
But things get most interesting when we add a coordinate:
da = xr.DataArray([9, 0, 2, 1, 0],
dims=['x'],
coords={'x': [10, 20, 30, 40, 50]})
da
Xarray has built-in plotting, like pandas.
da.plot(marker='o')
Multidimensional DataArray¶
If we are just dealing with 1D data, Pandas and Xarray have very similar capabilities. Xarray's real potential comes with multidimensional data.
Let's go back to the multidimensional ARGO data we loaded in the numpy lession. If you haven't already downloaded it, you can do so at the command line with
curl -O http://www.ldeo.columbia.edu/~rpa/argo_float_4901412.npz
We reload this data and examine its keys.
argo_data = np.load('argo_float_4901412.npz')
argo_data.keys()
The values of the argo_data object are numpy arrays.
S = argo_data.f.S
T = argo_data.f.T
P = argo_data.f.P
levels = argo_data.f.levels
lon = argo_data.f.lon
lat = argo_data.f.lat
date = argo_data.f.date
print(S.shape, lon.shape, date.shape)
Let's organize the data and coordinates of the salinity variable into a DataArray.
da_salinity = xr.DataArray(S, dims=['level', 'date'],
coords={'level': levels,
'date': date},)
da_salinity
da_salinity.plot(yincrease=False)
Attributes can be used to store metadata. What metadata should you store? The CF Conventions are a great resource for thinking about climate metadata. Below we define two of the required CF-conventions attributes.
da_salinity.attrs['units'] = 'PSU'
da_salinity.attrs['standard_name'] = 'sea_water_salinity'
da_salinity
Datasets¶
A Dataset holds many DataArrays which potentially can share coordinates. In analogy to pandas:
pandas.Series : pandas.Dataframe :: xarray.DataArray : xarray.Dataset
Constructing Datasets manually is a bit more involved in terms of syntax. The Dataset constructor takes three arguments:
data_varsshould be a dictionary with each key as the name of the variable and each value as one of:- A
DataArrayor Variable - A tuple of the form
(dims, data[, attrs]), which is converted into arguments for Variable - A pandas object, which is converted into a
DataArray - A 1D array or list, which is interpreted as values for a one dimensional coordinate variable along the same dimension as it’s name
- A
coordsshould be a dictionary of the same form as data_vars.attrsshould be a dictionary.
Let's put together a Dataset with temperature, salinity and pressure all together
argo = xr.Dataset(
data_vars={'salinity': (('level', 'date'), S),
'temperature': (('level', 'date'), T),
'pressure': (('level', 'date'), P)},
coords={'level': levels,
'date': date})
argo
What about lon and lat? We forgot them in the creation process, but we can add them after the fact.
argo['lon'] = lon
argo
That was not quite right...we want lon to have dimension date:
del argo['lon']
argo['lon'] = ('date', lon)
argo['lat'] = ('date', lat)
argo
Coordinates vs. Data Variables¶
Data variables can be modified through arithmentic operations or other functions. Coordinates are always keept the same.
argo * 10000
Clearly lon and lat are coordinates rather than data variables. We can change their status as follows:
argo = argo.set_coords(['lon', 'lat'])
argo
The * symbol in the representation above indicates that level and date are "dimension coordinates" (they describe the coordinates associated with data variable axes) while lon and lat are "non-dimension coordinates". We can make any variable a non-dimension coordiante.
argo.salinity[2].plot()
argo.salinity[:, 10].plot()
However, it is often much more powerful to use xarray's .sel() method to use label-based indexing.
argo.salinity.sel(level=2).plot()
argo.salinity.sel(date='2012-10-22').plot()
.sel() also supports slicing. Unfortunately we have to use a somewhat awkward syntax, but it still works.
argo.salinity.sel(date=slice('2012-10-01', '2012-12-01')).plot()
.sel() also works on the whole Dataset
argo.sel(date='2012-10-22')
Computation¶
Xarray dataarrays and datasets work seamlessly with arithmetic operators and numpy array functions.
α = np.cos(argo.temperature) * np.sin(argo.salinity)**2
α.plot()
Broadcasting¶
Broadcasting arrays in numpy is a nightmare. It is much easier when the data axes are labeled!
This is a useless calculation, but it illustrates how perfoming an operation on arrays with differenty coordinates will result in automatic broadcasting
level_times_lat = argo.level * argo.lat
level_times_lat.plot()
Reductions¶
Rather than performing reductions on axes (as in numpy), we can perform them on dimensions.
argo_mean = argo.mean(dim='date')
argo_mean
argo_mean.salinity.plot()
Aside: Swapping Dims¶
Now we can fix a pesky problem with this dataset: the fact that it uses level (rather than pressure) as the vertical coordinate
argo['pres_mean'] = argo_mean.pressure
argo_pcoords = argo.swap_dims({'level': 'pres_mean'})
argo_pcoords.salinity.plot(yincrease=False)
Groupby: Example with SST Climatology¶
Here will we work with SST data from NOAA's NERSST project. Download it by running
curl -O http://ldeo.columbia.edu/~rpa/NOAA_NCDC_ERSST_v3b_SST.nc
ds = xr.open_dataset('NOAA_NCDC_ERSST_v3b_SST.nc')
ds
ls
sst = ds.sst
sst.mean(dim='time').plot(vmin=-2, vmax=30)
sst.mean(dim=('time', 'lon')).plot()
sst_zonal_time_mean = sst.mean(dim=('time', 'lon'))
(sst.mean(dim='lon') - sst_zonal_time_mean).T.plot()
#sst.sel(lon=200, lat=0).plot()
sst.sel(lon=230, lat=0, method='nearest').plot()
sst.sel(lon=230, lat=45, method='nearest').plot()
# climatologies
sst_clim = sst.groupby('time.month').mean(dim='time')
sst_clim.sel(lon=230, lat=45, method='nearest').plot()
sst_clim.mean(dim='lon').T.plot.contourf(levels=np.arange(-2,30))
sst_anom = sst.groupby('time.month') - sst_clim
sst_anom.sel(lon=230, lat=45, method='nearest').plot()
sst_anom.std(dim='time').plot()
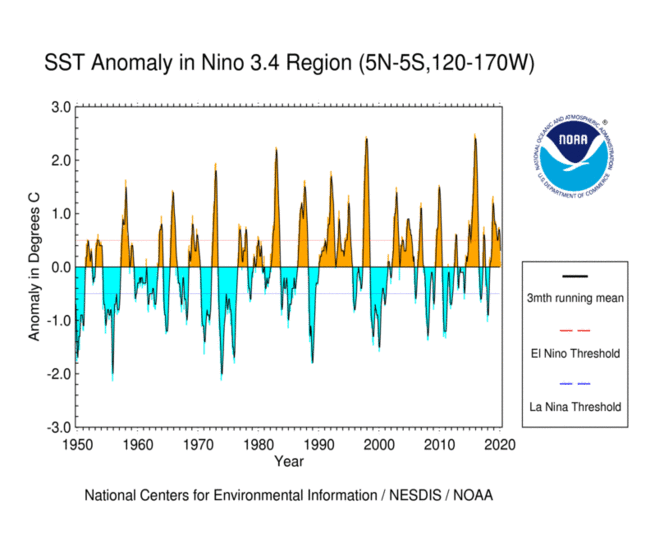
https://www.ncdc.noaa.gov/teleconnections/enso/indicators/sst.php
El Niño (La Niña) is a phenomenon in the equatorial Pacific Ocean characterized by a five consecutive 3-month running mean of sea surface temperature (SST) anomalies in the Niño 3.4 region that is above (below) the threshold of +0.5°C (-0.5°C). This standard of measure is known as the Oceanic Niño Index (ONI).
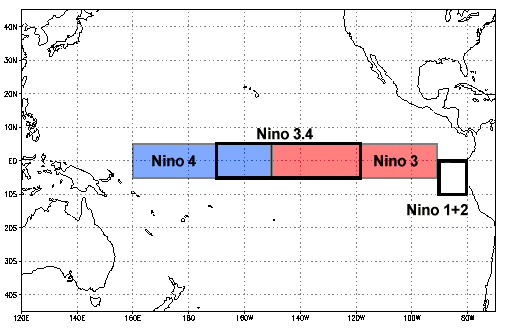
sst_anom_nino34 = sst_anom.sel(lat=slice(-5, 5), lon=slice(190, 240))
sst_anom_nino34[0].plot()
sst_anom_nino34_mean = sst_anom_nino34.mean(dim=('lon', 'lat'))
oni = sst_anom_nino34_mean.rolling(time=3).mean(dim='time')
fig, ax = plt.subplots()
sst_anom_nino34_mean.plot(ax=ax, label='raw')
oni.plot(ax=ax, label='smoothed')
ax.grid()
# create a categorical dataarray
nino34 = xr.full_like(oni, 'none', dtype='U4')
nino34[oni >= 0.5] = 'nino'
nino34[oni <= -0.5] = 'nina'
nino34
sst_nino_composite = sst_anom.groupby(nino34.rename('nino34')).mean(dim='time')
sst_nino_composite.sel(nino34='nino').plot()
sst_nino_composite.sel(nino34='nina').plot()
nino_ds = xr.Dataset({'nino34': nino34, 'oni': oni}).drop('month')
nino_ds.to_netcdf('nino34_index.nc')